Interpolation from 2D sem mesh
Our interpolation routines work mostly for 3D fields, therefore here we show a workaround by converting 2D fields into 3D fields with one element
Import general modules
[1]:
# Import required modules
from mpi4py import MPI #equivalent to the use of MPI_init() in C
import matplotlib.pyplot as plt
import numpy as np
# Get mpi info
comm = MPI.COMM_WORLD
Import modules from pysemtools
[2]:
from pysemtools.io.ppymech.neksuite import pynekread
from pysemtools.datatypes.msh import Mesh
from pysemtools.datatypes.coef import Coef
from pysemtools.datatypes.field import FieldRegistry
Read the data and build objects
In this instance, we create connectivity for the mesh object, given that we wish to use direct stiffness summation to reduce discontinuities.
[3]:
msh = Mesh(comm, create_connectivity=True)
fld = FieldRegistry(comm)
pynekread('../data/mixlay0.f00001', comm, data_dtype=np.double, msh=msh, fld=fld)
coef = Coef(msh, comm, get_area=False)
2024-08-26 08:37:50,343 - Mesh - INFO - Initializing empty Mesh object.
2024-08-26 08:37:50,344 - Field - INFO - Initializing empty Field object
2024-08-26 08:37:50,345 - pynekread - INFO - Reading file: ../data/mixlay0.f00001
2024-08-26 08:37:50,359 - Mesh - INFO - Initializing Mesh object from x,y,z ndarrays.
2024-08-26 08:37:50,360 - Mesh - INFO - Initializing common attributes.
2024-08-26 08:37:50,361 - Mesh - INFO - Creating connectivity
2024-08-26 08:37:50,545 - Mesh - INFO - Mesh object initialized.
2024-08-26 08:37:50,545 - Mesh - INFO - Mesh data is of type: float64
2024-08-26 08:37:50,546 - Mesh - INFO - Elapsed time: 0.18645260199999994s
2024-08-26 08:37:50,546 - pynekread - INFO - Reading field data
2024-08-26 08:37:50,553 - pynekread - INFO - File read
2024-08-26 08:37:50,554 - pynekread - INFO - Elapsed time: 0.209186977s
2024-08-26 08:37:50,555 - Coef - INFO - Initializing Coef object
2024-08-26 08:37:50,555 - Coef - INFO - Getting derivative matrices
2024-08-26 08:37:50,557 - Coef - INFO - Calculating the components of the jacobian
2024-08-26 08:37:50,565 - Coef - INFO - Calculating the jacobian determinant and inverse of the jacobian matrix
2024-08-26 08:37:50,569 - Coef - INFO - Calculating the mass matrix
2024-08-26 08:37:50,570 - Coef - INFO - Coef object initialized
2024-08-26 08:37:50,570 - Coef - INFO - Coef data is of type: float64
2024-08-26 08:37:50,571 - Coef - INFO - Elapsed time: 0.016067732s
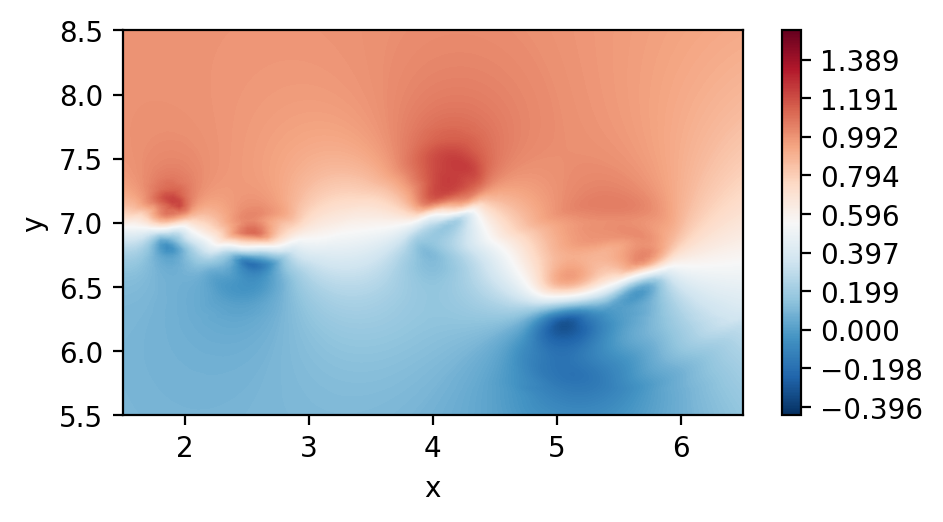
Plot the 2D velocity field
[4]:
fig, ax = plt.subplots(figsize=(5, 2.5), dpi = 200)
c = ax.tricontourf(msh.x.flatten(), msh.y.flatten() ,fld.registry["u"].flatten(), levels=100, cmap="RdBu_r")
fig.colorbar(c)
ax.set_aspect('equal')
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_xlim([1.5,6.5])
ax.set_ylim([5.5,8.5])
plt.show()
Extrude the mesh to be one element deep
For this we can simply use a helper function
[5]:
# Import helper function
from pysemtools.datatypes.utils import extrude_2d_sem_mesh
# Extrude the 2D mesh to 3D
msh3d, fld3d = extrude_2d_sem_mesh(comm, lz = msh.lx, msh = msh, fld = fld)
2024-08-26 08:37:51,889 - Mesh - INFO - Initializing Mesh object from x,y,z ndarrays.
2024-08-26 08:37:51,890 - Mesh - INFO - Initializing common attributes.
2024-08-26 08:37:51,891 - Mesh - INFO - Creating connectivity
2024-08-26 08:37:52,615 - Mesh - INFO - Mesh object initialized.
2024-08-26 08:37:52,616 - Mesh - INFO - Mesh data is of type: float64
2024-08-26 08:37:52,617 - Mesh - INFO - Elapsed time: 0.7274203159999999s
2024-08-26 08:37:52,618 - Field - INFO - Initializing empty Field object
Generate interpolation points
The interpolation points must have 3 coordinates, so we simply set the z limits to be zero, since in our extrudex 3d mesh, zero is part of the extruded dimension
[6]:
# Import helper functions
import pysemtools.interpolation.utils as interp_utils
import pysemtools.interpolation.pointclouds as pcs
if comm.Get_rank() == 0 :
# Generate the bounding box of the points
x_bbox = [1.5, 6.5]
y_bbox = [5.5, 8.5]
z_bbox = [0,0]
nx = 100
ny = 100
nz = 1
# Generate the 1D mesh
x_1d = pcs.generate_1d_arrays(x_bbox, nx, mode="equal")
y_1d = pcs.generate_1d_arrays(y_bbox, ny, mode="equal")
z_1d = pcs.generate_1d_arrays(z_bbox, nz, mode="equal")
# Generate a 3D mesh
x, y, z = np.meshgrid(x_1d, y_1d, z_1d, indexing='ij')
# Array the points as a list of probes
xyz = interp_utils.transform_from_array_to_list(nx,ny,nz,[x, y, z])
# Write the points for future use
with open('points.csv', 'w') as f:
for i in range((xyz.shape[0])):
f.write(f"{xyz[i][0]},{xyz[i][1]},{xyz[i][2]}\n")
else:
xyz = 1
Interpolate
From here on, the process is the same
Find the points
The points are always found at the initialization stage. This might be time consiming
[7]:
# Interpolate the interpolation module
from pysemtools.interpolation.probes import Probes
probes = Probes(comm, probes = xyz, msh = msh3d, point_interpolator_type='multiple_point_legendre_numpy', max_pts=256, find_points_comm_pattern='point_to_point')
2024-08-26 08:37:52,671 - Probes - INFO - Initializing Probes object
2024-08-26 08:37:52,672 - Probes - INFO - No input file provided. Using default values
2024-08-26 08:37:52,673 - Probes - INFO - Probes provided as keyword argument. Assigning in rank 0
2024-08-26 08:37:52,674 - Probes - INFO - Mesh provided as keyword argument
2024-08-26 08:37:52,674 - Probes - INFO - Initializing interpolator
2024-08-26 08:37:52,675 - Interpolator - INFO - Initializing Interpolator object
2024-08-26 08:37:52,675 - Interpolator - INFO - Initializing point interpolator: multiple_point_legendre_numpy
2024-08-26 08:37:52,680 - Interpolator - INFO - Allocating buffers in point interpolator
2024-08-26 08:37:52,681 - Interpolator - INFO - Using device: cpu
2024-08-26 08:37:52,682 - Interpolator - INFO - Interpolator initialized
2024-08-26 08:37:52,682 - Interpolator - INFO - Elapsed time: 0.006703410999999715s
2024-08-26 08:37:52,683 - Probes - INFO - Setting up global tree
2024-08-26 08:37:52,683 - Interpolator - INFO - Using global_tree of type: rank_bbox
2024-08-26 08:37:52,684 - Interpolator - INFO - Finding bounding boxes for each rank
2024-08-26 08:37:52,686 - Interpolator - INFO - Creating global KD tree with rank centroids
2024-08-26 08:37:52,688 - Interpolator - INFO - Elapsed time: 0.004152533000000069s
2024-08-26 08:37:52,688 - Probes - INFO - Scattering probes to all ranks
2024-08-26 08:37:52,689 - Interpolator - INFO - Scattering probes
2024-08-26 08:37:52,691 - Interpolator - INFO - done
2024-08-26 08:37:52,693 - Interpolator - INFO - Elapsed time: 0.001545761999999673s
2024-08-26 08:37:52,693 - Probes - INFO - Finding points
2024-08-26 08:37:52,694 - Interpolator - INFO - using communication pattern: point_to_point
2024-08-26 08:37:52,695 - Interpolator - INFO - Finding points - start
2024-08-26 08:37:52,696 - Interpolator - INFO - Finding bounding box of sem mesh
2024-08-26 08:37:52,728 - Interpolator - INFO - Creating KD tree with local bbox centroids
2024-08-26 08:37:52,729 - Interpolator - INFO - Obtaining candidate ranks and sources
2024-08-26 08:37:52,732 - Interpolator - INFO - Send data to candidates and recieve from sources
2024-08-26 08:37:52,733 - Interpolator - INFO - Find rst coordinates for the points
2024-08-26 08:37:53,891 - Interpolator - INFO - Send data to sources and recieve from candidates
2024-08-26 08:37:53,892 - Interpolator - INFO - Determine which points were found and find best candidate
2024-08-26 08:37:53,921 - Interpolator - INFO - Finding points - finished
2024-08-26 08:37:53,922 - Interpolator - INFO - Elapsed time: 1.2260808879999998s
2024-08-26 08:37:53,922 - Probes - INFO - Gathering probes to rank 0 after search
2024-08-26 08:37:53,923 - Interpolator - INFO - Gathering probes
2024-08-26 08:37:53,924 - Interpolator - INFO - done
2024-08-26 08:37:53,925 - Interpolator - INFO - Elapsed time: 0.0014982169999999684s
2024-08-26 08:37:53,927 - Probes - INFO - Redistributing probes to found owners
2024-08-26 08:37:53,929 - Probes - INFO - Writing probe coordinates to ./interpolated_fields.csv
2024-08-26 08:37:53,948 - Probes - INFO - Writing points with warnings to ./warning_points_interpolated_fields.json
2024-08-26 08:37:53,950 - Probes - INFO - Found 10000 points, 0 not found, 0 with warnings
2024-08-26 08:37:53,951 - Probes - INFO - Probes object initialized
2024-08-26 08:37:53,951 - Probes - INFO - Elapsed time: 1.2801558099999997s
Interpolate
After the points are found, any field can be interpolated
[8]:
# Interpolate from a field list
probes.interpolate_from_field_list(0, [fld3d.registry['u']], comm, write_data=False)
2024-08-26 08:37:53,960 - Probes - INFO - Interpolating fields from field list
2024-08-26 08:37:53,964 - Probes - INFO - Interpolating field 0
2024-08-26 08:37:53,965 - Interpolator - INFO - Interpolating field from rst coordinates
2024-08-26 08:37:54,002 - Interpolator - INFO - Elapsed time: 0.03556546699999963s
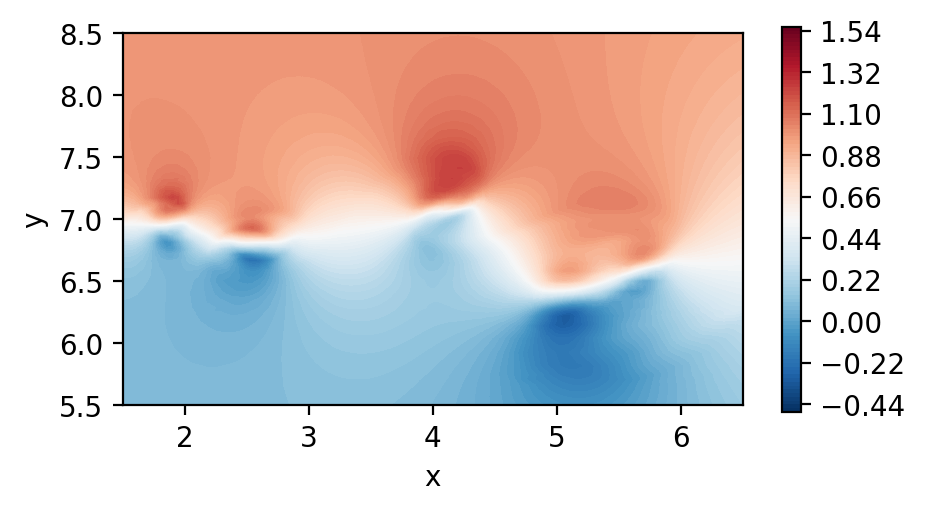
Process the results
In the current implementation, points are partitioned in ranks but rank zero also has them all collected, so we work there.
[9]:
if comm.Get_rank() == 0:
int_fields = interp_utils.transform_from_list_to_array(nx,ny,nz,probes.interpolated_fields)
u_int = int_fields[1]
#levels = 500
levels = np.linspace(-0.44, 1.54, 500)
cmapp='RdBu_r'
fig, ax = plt.subplots(1, 1, figsize=(5, 2.5), dpi = 200)
c1 = ax.tricontourf(x.flatten(), y.flatten() ,u_int.flatten(), levels=levels, cmap=cmapp)
fig.colorbar(c1)
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.show()