Interpolation to query points
In this example we show how to interpolate to an arbitrary set of query points.
Import general modules
[1]:
# Import required modules
from mpi4py import MPI #equivalent to the use of MPI_init() in C
import matplotlib.pyplot as plt
import numpy as np
# Get mpi info
comm = MPI.COMM_WORLD
Import modules from pynek
[2]:
from pysemtools.io.ppymech.neksuite import pynekread
from pysemtools.datatypes.msh import Mesh
from pysemtools.datatypes.field import FieldRegistry
Read the data and build objects
In this instance, we create connectivity for the mesh object, given that we wish to use direct stiffness summation to reduce discontinuities.
[3]:
msh = Mesh(comm, create_connectivity=True)
fld = FieldRegistry(comm)
fname = '../data/rbc0.f00001'
pynekread(fname, comm, data_dtype=np.double, msh=msh, fld=fld)
2024-09-11 18:27:03,861 - Mesh - INFO - Initializing empty Mesh object.
2024-09-11 18:27:03,863 - Field - INFO - Initializing empty Field object
2024-09-11 18:27:03,864 - pynekread - INFO - Reading file: ../data/rbc0.f00001
2024-09-11 18:27:03,876 - Mesh - INFO - Initializing Mesh object from x,y,z ndarrays.
2024-09-11 18:27:03,876 - Mesh - INFO - Initializing common attributes.
2024-09-11 18:27:03,877 - Mesh - INFO - Getting vertices
2024-09-11 18:27:03,878 - Mesh - INFO - Getting vertices
2024-09-11 18:27:03,888 - Mesh - INFO - Getting facet centers
2024-09-11 18:27:03,896 - Mesh - INFO - Creating connectivity
2024-09-11 18:27:04,376 - Mesh - INFO - Mesh object initialized.
2024-09-11 18:27:04,377 - Mesh - INFO - Mesh data is of type: float64
2024-09-11 18:27:04,378 - Mesh - INFO - Elapsed time: 0.502331771s
2024-09-11 18:27:04,378 - pynekread - INFO - Reading field data
2024-09-11 18:27:04,389 - pynekread - INFO - File read
2024-09-11 18:27:04,389 - pynekread - INFO - Elapsed time: 0.525439139s
Building the set of points to use
In this case, we will use a cylindrical mesh to interpolate the points in our domain.
For this, we will use some tools to generate point distributions.
Be mindful that in general, the interpolation routines will only takei nto considerations the points that are passed in rank 0.
For this reason, we build all the points only in this rank and generate a dummy variable xyz in other ranks.
[4]:
# Import helper functions
import pysemtools.interpolation.utils as interp_utils
import pysemtools.interpolation.pointclouds as pcs
if comm.Get_rank() == 0 :
# Generate the bounding box of the points
x_bbox = [0, 0.05]
y_bbox = [0, 2*np.pi]
z_bbox = [0 , 1]
nx = 30
ny = 30
nz = 80
# Generate the 1D mesh
x_1d = pcs.generate_1d_arrays(x_bbox, nx, mode="equal")
y_1d = pcs.generate_1d_arrays(y_bbox, ny, mode="equal")
z_1d = pcs.generate_1d_arrays(z_bbox, nz, mode="equal")
# Generate a 3D mesh
r, th, z = np.meshgrid(x_1d, y_1d, z_1d, indexing='ij')
x = r*np.cos(th)
y = r*np.sin(th)
# Array the points as a list of probes
xyz = interp_utils.transform_from_array_to_list(nx,ny,nz,[x, y, z])
# Write the points for future use
with open('points.csv', 'w') as f:
for i in range((xyz.shape[0])):
f.write(f"{xyz[i][0]},{xyz[i][1]},{xyz[i][2]}\n")
else:
xyz = 1
Interpolate
The module to use for the interpolations is:
[5]:
from pysemtools.interpolation.probes import Probes
Interpolate from points in memory
We have created the xyz points and we have them in memory, so we can just interpolate form there.
Finding the points
The first step is to initialize the probe object, which will attempt to find the query points in the SEM mesh.
There are many options to do this, so we recommend that you check the documentation for this class.
[6]:
probes = Probes(comm, probes = xyz, msh = msh, point_interpolator_type='multiple_point_legendre_numpy', max_pts=256, find_points_comm_pattern='point_to_point')
2024-09-11 18:27:05,085 - Probes - INFO - Initializing Probes object
2024-09-11 18:27:05,087 - Probes - INFO - Probes provided as keyword argument
2024-09-11 18:27:05,089 - Probes - INFO - Input probes are distributed: False
2024-09-11 18:27:05,090 - Probes - INFO - Mesh provided as keyword argument
2024-09-11 18:27:05,096 - Probes - INFO - Initializing interpolator
2024-09-11 18:27:05,098 - Interpolator - INFO - Initializing Interpolator object
2024-09-11 18:27:05,105 - Interpolator - INFO - Initializing point interpolator: multiple_point_legendre_numpy
2024-09-11 18:27:05,118 - Interpolator - INFO - Allocating buffers in point interpolator
2024-09-11 18:27:05,121 - Interpolator - INFO - Using device: cpu
2024-09-11 18:27:05,126 - Interpolator - INFO - Interpolator initialized
2024-09-11 18:27:05,127 - Interpolator - INFO - Elapsed time: 0.02208092599999989s
2024-09-11 18:27:05,128 - Probes - INFO - Setting up global tree
2024-09-11 18:27:05,129 - Interpolator - INFO - Using global_tree of type: rank_bbox
2024-09-11 18:27:05,133 - Interpolator - INFO - Finding bounding boxes for each rank
2024-09-11 18:27:05,136 - Interpolator - INFO - Creating global KD tree with rank centroids
2024-09-11 18:27:05,137 - Interpolator - INFO - Elapsed time: 0.004132310999999778s
2024-09-11 18:27:05,139 - Probes - INFO - Scattering probes to all ranks
2024-09-11 18:27:05,140 - Interpolator - INFO - Scattering probes
2024-09-11 18:27:05,157 - Interpolator - INFO - done
2024-09-11 18:27:05,159 - Interpolator - INFO - Elapsed time: 0.017513389999999962s
2024-09-11 18:27:05,159 - Probes - INFO - Finding points
2024-09-11 18:27:05,160 - Interpolator - INFO - using communication pattern: point_to_point
2024-09-11 18:27:05,164 - Interpolator - INFO - Finding points - start
2024-09-11 18:27:05,165 - Interpolator - INFO - Finding bounding box of sem mesh
2024-09-11 18:27:05,223 - Interpolator - INFO - Creating KD tree with local bbox centroids
2024-09-11 18:27:05,235 - Interpolator - INFO - Obtaining candidate ranks and sources
2024-09-11 18:27:05,376 - Interpolator - INFO - Send data to candidates and recieve from sources
2024-09-11 18:27:05,382 - Interpolator - INFO - Find rst coordinates for the points
2024-09-11 18:27:25,039 - Interpolator - INFO - Send data to sources and recieve from candidates
2024-09-11 18:27:25,042 - Interpolator - INFO - Determine which points were found and find best candidate
2024-09-11 18:27:25,406 - Interpolator - INFO - Finding points - finished
2024-09-11 18:27:25,408 - Interpolator - INFO - Elapsed time: 20.242600142s
2024-09-11 18:27:25,408 - Probes - INFO - Gathering probes to rank 0 after search
2024-09-11 18:27:25,409 - Interpolator - INFO - Gathering probes
2024-09-11 18:27:25,417 - Interpolator - INFO - done
2024-09-11 18:27:25,418 - Interpolator - INFO - Elapsed time: 0.007374879000000334s
2024-09-11 18:27:25,418 - Probes - INFO - Redistributing probes to found owners
2024-09-11 18:27:25,419 - Interpolator - INFO - Scattering probes
2024-09-11 18:27:25,427 - Interpolator - INFO - done
2024-09-11 18:27:25,428 - Interpolator - INFO - Elapsed time: 0.008342505000001665s
2024-09-11 18:27:25,429 - Probes - INFO - Writing probe coordinates to ./interpolated_fields.csv
2024-09-11 18:27:26,224 - Probes - INFO - Writing points with warnings to ./warning_points_interpolated_fields.json
2024-09-11 18:27:26,226 - Probes - INFO - Found 69760 points, 0 not found, 2240 with warnings
2024-09-11 18:27:26,227 - Probes - WARNING - There are points with warnings. Check the warning file to see them (error code -10)
2024-09-11 18:27:26,228 - Probes - WARNING - There are points with warnings. If test pattern is small, you can trust the interpolation
2024-09-11 18:27:26,229 - Probes - INFO - Probes object initialized
2024-09-11 18:27:26,230 - Probes - INFO - Elapsed time: 21.144440734s
To interpolate a particular field, you must call the function and put the fields in a list, as follow:
[7]:
# The first input is the time to write in the file
probes.interpolate_from_field_list(0, [fld.registry['w']], comm, write_data=False)
2024-09-11 18:27:26,237 - Probes - INFO - Interpolating fields from field list
2024-09-11 18:27:26,239 - Probes - INFO - Interpolating field 0
2024-09-11 18:27:26,239 - Interpolator - INFO - Interpolating field from rst coordinates
2024-09-11 18:27:26,660 - Interpolator - INFO - Elapsed time: 0.42095929099999907s
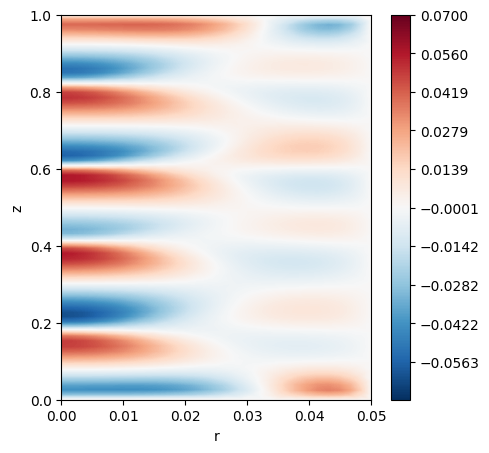
The points have now been interpolated and are gathered once again in rank 0. Therefore one can proceed to perform operations such as plotting.
Note that they are still arrayed as a list. If one whishes to use the structured mesh format, then the points must be mapped back to their original shape.
[8]:
if comm.Get_rank() == 0:
polar_fields = interp_utils.transform_from_list_to_array(nx,ny,nz,probes.interpolated_fields)
w_polar = polar_fields[1]
w_2d = np.mean(w_polar, axis=1)
levels = 500
levels = np.linspace(-0.07, 0.07, levels)
cmapp='RdBu_r'
fig, ax = plt.subplots(1, 1,figsize=(5, 5))
c1 = ax.tricontourf(r[:,0,:].flatten(), z[:,0,:].flatten() ,w_2d.flatten(), levels=levels, cmap=cmapp)
fig.colorbar(c1)
ax.set_xlabel("r")
ax.set_ylabel("z")
plt.show()
If the points are already in a csv file, we can choose to interpolate from those, instead of bulding points in memory (which we already did).
One option is to of course read them and assign them to the xyz array we indicated before. The same can be done with the mesh.
Simply write the path to the file where the mesh and/or the probes are in the appropiate keyword argument.
Initializing the probes object
Now we can initialize the probes object
[9]:
probes2 = Probes(comm, probes="./points.csv", msh = '../data/rbc0.f00001', point_interpolator_type='multiple_point_legendre_numpy', max_pts=256, find_points_comm_pattern='point_to_point')
2024-09-11 18:27:27,207 - Probes - INFO - Initializing Probes object
2024-09-11 18:27:27,208 - Probes - INFO - Reading probes from ./points.csv
2024-09-11 18:27:27,388 - Probes - INFO - Input probes are distributed: False
2024-09-11 18:27:27,388 - Probes - INFO - Reading mesh from ../data/rbc0.f00001
2024-09-11 18:27:27,389 - Mesh - INFO - Initializing empty Mesh object.
2024-09-11 18:27:27,390 - pynekread - INFO - Reading file: ../data/rbc0.f00001
2024-09-11 18:27:27,393 - Mesh - INFO - Initializing Mesh object from x,y,z ndarrays.
2024-09-11 18:27:27,394 - Mesh - INFO - Initializing common attributes.
2024-09-11 18:27:27,394 - Mesh - INFO - Getting vertices
2024-09-11 18:27:27,395 - Mesh - INFO - Getting vertices
2024-09-11 18:27:27,403 - Mesh - INFO - Getting facet centers
2024-09-11 18:27:27,409 - Mesh - INFO - Mesh object initialized.
2024-09-11 18:27:27,410 - Mesh - INFO - Mesh data is of type: float32
2024-09-11 18:27:27,410 - Mesh - INFO - Elapsed time: 0.016976128999999673s
2024-09-11 18:27:27,411 - pynekread - INFO - File read
2024-09-11 18:27:27,412 - pynekread - INFO - Elapsed time: 0.022214138999999022s
2024-09-11 18:27:27,412 - Probes - INFO - Initializing interpolator
2024-09-11 18:27:27,413 - Interpolator - INFO - Initializing Interpolator object
2024-09-11 18:27:27,413 - Interpolator - INFO - Initializing point interpolator: multiple_point_legendre_numpy
2024-09-11 18:27:27,418 - Interpolator - INFO - Allocating buffers in point interpolator
2024-09-11 18:27:27,418 - Interpolator - INFO - Using device: cpu
2024-09-11 18:27:27,419 - Interpolator - INFO - Interpolator initialized
2024-09-11 18:27:27,420 - Interpolator - INFO - Elapsed time: 0.0066051529999988645s
2024-09-11 18:27:27,421 - Probes - INFO - Setting up global tree
2024-09-11 18:27:27,421 - Interpolator - INFO - Using global_tree of type: rank_bbox
2024-09-11 18:27:27,422 - Interpolator - INFO - Finding bounding boxes for each rank
2024-09-11 18:27:27,424 - Interpolator - INFO - Creating global KD tree with rank centroids
2024-09-11 18:27:27,424 - Interpolator - INFO - Elapsed time: 0.002681530000000265s
2024-09-11 18:27:27,425 - Probes - INFO - Scattering probes to all ranks
2024-09-11 18:27:27,425 - Interpolator - INFO - Scattering probes
2024-09-11 18:27:27,434 - Interpolator - INFO - done
2024-09-11 18:27:27,435 - Interpolator - INFO - Elapsed time: 0.008592331000002673s
2024-09-11 18:27:27,435 - Probes - INFO - Finding points
2024-09-11 18:27:27,436 - Interpolator - INFO - using communication pattern: point_to_point
2024-09-11 18:27:27,436 - Interpolator - INFO - Finding points - start
2024-09-11 18:27:27,437 - Interpolator - INFO - Finding bounding box of sem mesh
2024-09-11 18:27:27,467 - Interpolator - INFO - Creating KD tree with local bbox centroids
2024-09-11 18:27:27,476 - Interpolator - INFO - Obtaining candidate ranks and sources
2024-09-11 18:27:27,597 - Interpolator - INFO - Send data to candidates and recieve from sources
2024-09-11 18:27:27,602 - Interpolator - INFO - Find rst coordinates for the points
2024-09-11 18:27:45,167 - Interpolator - INFO - Send data to sources and recieve from candidates
2024-09-11 18:27:45,170 - Interpolator - INFO - Determine which points were found and find best candidate
2024-09-11 18:27:45,529 - Interpolator - INFO - Finding points - finished
2024-09-11 18:27:45,530 - Interpolator - INFO - Elapsed time: 18.093211668s
2024-09-11 18:27:45,530 - Probes - INFO - Gathering probes to rank 0 after search
2024-09-11 18:27:45,531 - Interpolator - INFO - Gathering probes
2024-09-11 18:27:45,536 - Interpolator - INFO - done
2024-09-11 18:27:45,536 - Interpolator - INFO - Elapsed time: 0.00425026900000347s
2024-09-11 18:27:45,537 - Probes - INFO - Redistributing probes to found owners
2024-09-11 18:27:45,537 - Interpolator - INFO - Scattering probes
2024-09-11 18:27:45,543 - Interpolator - INFO - done
2024-09-11 18:27:45,544 - Interpolator - INFO - Elapsed time: 0.005580473000001973s
2024-09-11 18:27:45,544 - Probes - INFO - Writing probe coordinates to ./interpolated_fields.csv
2024-09-11 18:27:46,229 - Probes - INFO - Writing points with warnings to ./warning_points_interpolated_fields.json
2024-09-11 18:27:46,232 - Probes - INFO - Found 69760 points, 0 not found, 2240 with warnings
2024-09-11 18:27:46,233 - Probes - WARNING - There are points with warnings. Check the warning file to see them (error code -10)
2024-09-11 18:27:46,234 - Probes - WARNING - There are points with warnings. If test pattern is small, you can trust the interpolation
2024-09-11 18:27:46,235 - Probes - INFO - Probes object initialized
2024-09-11 18:27:46,235 - Probes - INFO - Elapsed time: 19.027900337s
After this, you can simply follow the same steps to interpolate fields. You can write the fields to the same data type by using the appropiate command.
[10]:
probes2.interpolate_from_field_list(0, [fld.registry['w']], comm, write_data=True)
2024-09-11 18:27:46,242 - Probes - INFO - Interpolating fields from field list
2024-09-11 18:27:46,244 - Probes - INFO - Interpolating field 0
2024-09-11 18:27:46,245 - Interpolator - INFO - Interpolating field from rst coordinates
2024-09-11 18:27:46,703 - Interpolator - INFO - Elapsed time: 0.4582567080000004s
2024-09-11 18:27:46,706 - Probes - INFO - Writing interpolated fields to ./interpolated_fields.csv
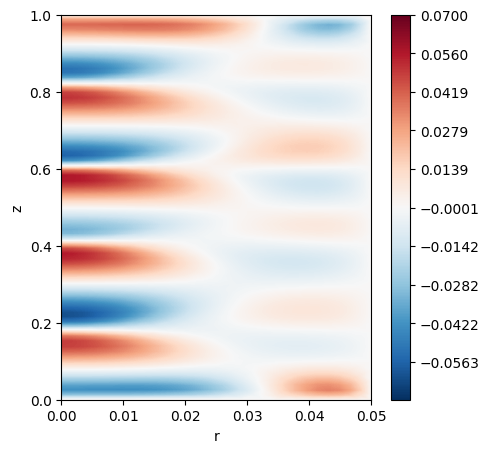
Reading probes results from CSV
To read probes, we can use the probe reader object. Note that we will do this in rank 0. This is also the format of Neko probes, therefore you will be able to read those too.
[11]:
# Import the module
from pysemtools.io.read_probes import ProbesReader
if comm.Get_rank() == 0:
pr = ProbesReader(file_name = './interpolated_fields.csv')
# The name of the field is the index in the list when interpolated by default
w_polar2 = interp_utils.transform_from_list_to_array(nx,ny,nz,pr.fields['0'])
w_2d2 = np.mean(w_polar2[0], axis=1)
levels = 500
levels = np.linspace(-0.07, 0.07, levels)
cmapp='RdBu_r'
fig, ax = plt.subplots(1, 1,figsize=(5, 5))
c1 = ax.tricontourf(r[:,0,:].flatten(), z[:,0,:].flatten() ,w_2d2.flatten(), levels=levels, cmap=cmapp)
fig.colorbar(c1)
ax.set_xlabel("r")
ax.set_ylabel("z")
plt.show()